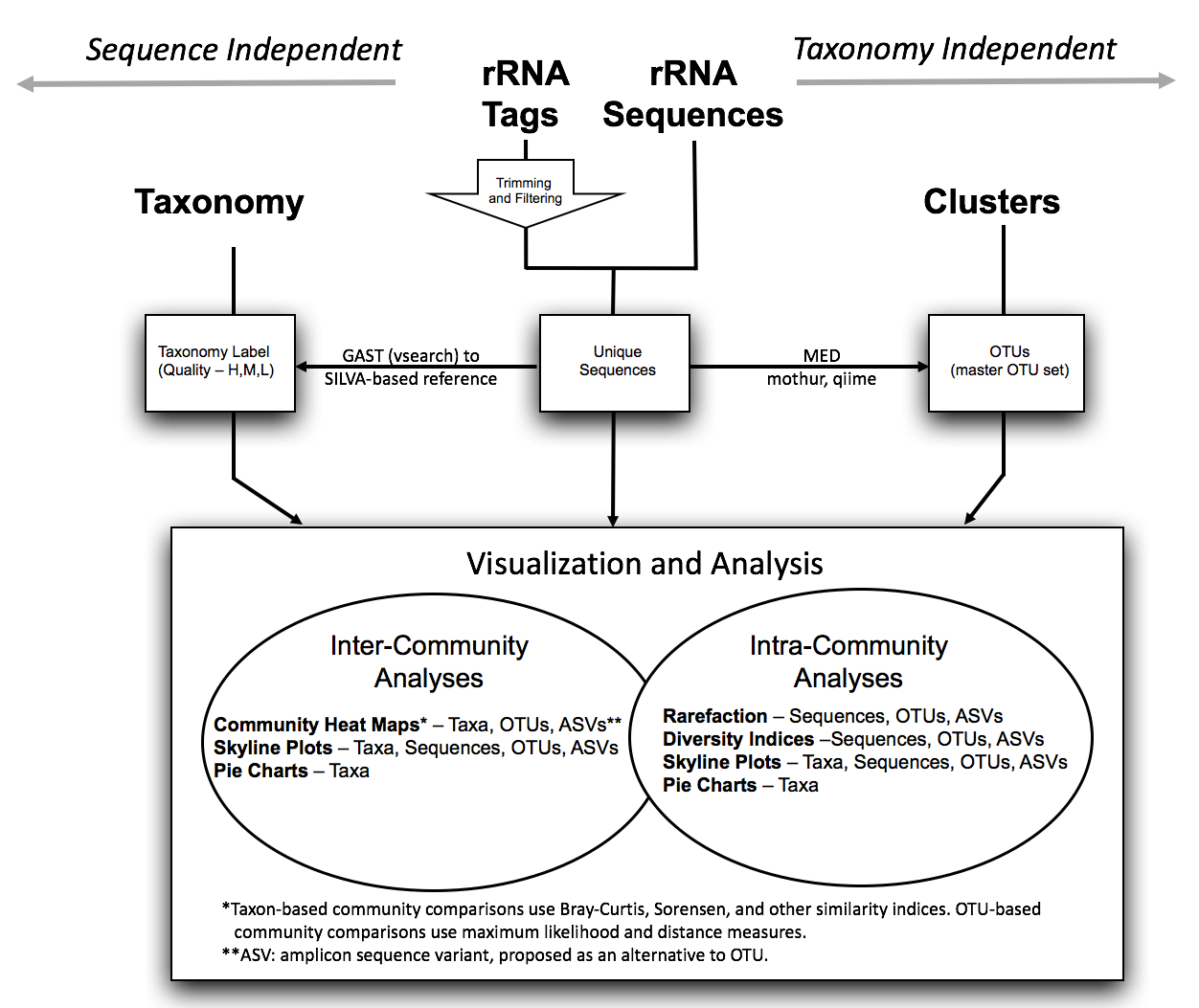
Overview
- The Visualization and Analysis of Microbial Population Structures project is an integrated collection of tools with which researchers can visualize and analyze data for microbial population structures and distributions. The project includes visualization tools such as heat maps that simultaneously compare the taxonomic distributions of multiple datasets and 3-D charts of the frequency distributions of 16S rRNA tags. Analytical tools include Chao diversity estimates and rarefaction curves. As a service to the community, researchers have the opportunity to upload their own data to the site for private viewing with the full range of data and analysis tools. Public data can be downloaded for further analysis locally.
- Background: The phylogenetic and phenotypic diversity of microorganisms eclipses the genetic and biochemical repertoire of the plants and animals combined. Until very recently, sophisticated statistical approaches borrowed from macro-ecology have been sufficient to measure population structure differences between microbial communities. However, the recent introduction of massively-parallel, high-throughput DNA sequencing technologies, e.g. 454 pyrosequencing, enables analyses that were previously considered impractical because of the required sampling intensity and complex experimental designs. To harness the power of these information-rich datasets, it is necessary to develop new analytical techniques and tools for describing and comparing microbial communities.
- Objectives: VAMPS is a publicly-accessible website to measure and visualize similarities and differences between molecular profiles of complex microbial communities. The primary objective of VAMPS is to improve the statistical metrics and to visualize computed similarities and differences between multiple, molecular-based microbial community profiles. VAMPS is initially focusing on analyses of massively large data sets of short, hypervariable sequences of rRNA genes (V6-rRNA tags) from naturally occurring microbial populations. The VAMPS tools will extend to community-based comparisons of full-length ribosomal RNAs (rRNAs) and, ultimately, to metagenomic (the collective sequence of genomes in a mixed microbial community) data. In the future, VAMPS tools will allow microbial ecologists to integrate population structure information with geochemical, physical and other contextual data at sites of interest.